This post was written by Janina Lisa Kypke.
 You might be wondering what animals am I talking about and how large is the largest animal family? Meet the rove beetles or Staphylinidae!
You might be wondering what animals am I talking about and how large is the largest animal family? Meet the rove beetles or Staphylinidae!
- Rove beetles consist of more than 63.137 fossil and living species. This is the last official number published in 2017, which means it is already outdated by now since more species are being described every year.
- Rove beetles can be found on every continent except Antarctica, their preferred habitat is shady and moist as can be found e.g. in leaf litter, at shores or in the dung. They vary in size and can be as small as only 0.5 mm and up to 5 cm long.
- Most of the rove beetles are easily identifiable because their hind wings are shortened and wings are folded underneath. This makes them much more flexible and faster than other beetles and might be why they are such efficient and numerous predators.
Oh, and Jurassic Park was wrong. Unfortunately we are not able to extract DNA from fossils, not even from comparatively young amber fossils. Sorry to burst that bubble, I know it would have been amazing.
At this point I am very happy to tell you that the first Staphylinid phylogeny based on (mostly) genomic data is about to be finished. This work was done in a collaborative effort with members of the 1KITE team, CSIRO, and the Bioinformatics Institute in St Petersburg where we managed to obtain close to 4000 genes for 33 transcriptomes and genomes across all Staphylinoidea. More taxa with fewer genes that have already been published are added to have a more complete representation of the group.
Getting these results is super exciting since they will shed light on the higher-level relationships amongst the subfamilies and groups of subfamilies. Among the answers I am hoping to find are for example whether there are indeed four distinct groups of subfamilies as morphology suggests or which subfamilies are the oldest/ youngest from an evolutionary point of view. These results can help us to begin to understand how this family of beetles could become so large and omnipresent and successful.
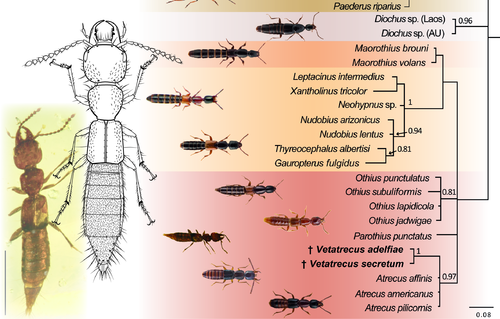
Rove beetle amber fossil with corresponding line drawing and its placement in a phylogenetic tree
During my research I was able to expand my background in ecology not only to bioinformatics for phylogenomics but I also had the chance to study amber fossils both using traditional light microscopy as well as X-ray computed microtomography (µCT) and I described my first species!
BIG4 have undoubtedly expanded my scientific and private network. During the meetings we were able to freely discuss our research, share ideas and make plans for the future. Since the network is interdisciplinary, I got the chance to expand my knowledge not only in my field of research but also beyond. I am very grateful to have been part of the BIG4 and cannot wait to see how we will continue to shape the future.
Publications within Janina's project:
- J.L. Kypke, A. Solodovnikov, A. Brunke, S. Yamamoto, D. Zyla (2018) The past and the present through phylogenetic analysis: the rove beetle tribe Othiini now and 99 Ma, Systematic Entomology. DOI:10.1111/syen.12305.
- First phylogeny for Staphylinidae based on genomic data. (In preparation)
- X-ray µ-CT helps to identify first fossil Orsunius sp. in Baltic amber. (In preparation)
- Baltic amber fossils reveal an ancient connection between disjunct distribution of recent species. (In preparation)

